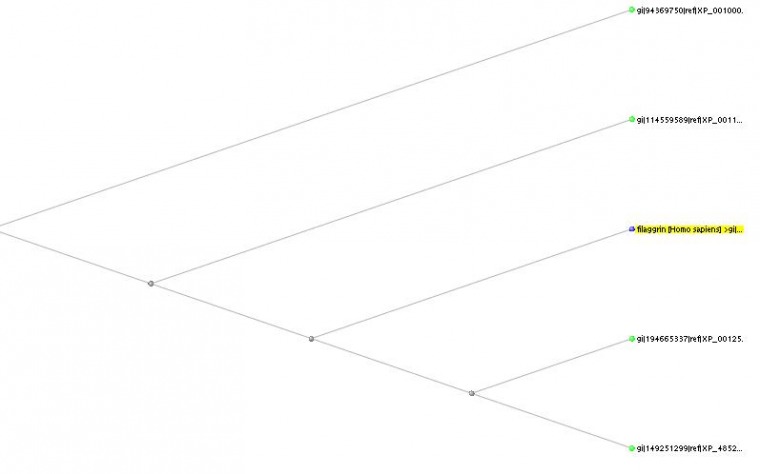
Homologues of the Filaggrin protein
Lookng at the homologies of the protein sequences, I once again found that the Chimpanzee had the highest similarity match. Again, a E-value of zero indicates a nearly identical match, while higher values show higher likelihood of coincidence.
Pan troglodytes (Chimpanzee)
Name: PREDICTED: filaggrin isoform 1 [Pan troglodytes]
Maximum Identification: 100%
Length: 3088 aa
Accession number: XM_001134714.1
GI: 114559589
E-value: 0.0
Bos taurus (Cow)
Name: PREDICTED: similar to filaggrin [Bos taurus]
Maximum Identification: 99%
Length: 1656 aa
Accession number: XM_001255582.2
GI: 194665337
E-value: 3e-154
Mus musculus (Mouse)
Name: PREDICTED: filaggrin [Mus musculus]
Maximum Identification: 22%
Length: 3541 aa
Accession number: XM_485270.5
GI: 149251299
E-value: 7e-09
Mus musculus (Mouse)
Name: PREDICTED: similar to filaggrin [Mus musculus]
Maximum Identification: 40%
Length: 987 aa
Accession number: XM_001000758.1
GI: 94369750
E-value: 8e-08
Analysis
This time, a surprising result was seen in the Filaggrin homologue in the Bos taurus. While it showed a low similarity in the alignment of the mRNA sequences, it showed a 99% similarity in the protein sequence when aligned against the Homo sapien sequence using BLAST.
What this indicates is that although many times it is seen that the gene sequence shows a higher similarity between homologues due to conservation that is later lost after splicing, our gene requires a highly conserved end result, as displayed in the in highly conserved protein result seen in this BLAST, at least in this cow homologue.
In my attempt to use ClustalW, T-COFFEE, and Muscle alignment programs, I was unsuccessful due to the length of my sequences. While these are powerful alignment programs, their use becomes diminished when unable to be utilized with any sequence that exceeds this unfortunate limitation.
This time, a surprising result was seen in the Filaggrin homologue in the Bos taurus. While it showed a low similarity in the alignment of the mRNA sequences, it showed a 99% similarity in the protein sequence when aligned against the Homo sapien sequence using BLAST.
What this indicates is that although many times it is seen that the gene sequence shows a higher similarity between homologues due to conservation that is later lost after splicing, our gene requires a highly conserved end result, as displayed in the in highly conserved protein result seen in this BLAST, at least in this cow homologue.
In my attempt to use ClustalW, T-COFFEE, and Muscle alignment programs, I was unsuccessful due to the length of my sequences. While these are powerful alignment programs, their use becomes diminished when unable to be utilized with any sequence that exceeds this unfortunate limitation.
As we saw in then mRNA homologies, there were two homologues in mice. While looking at the protein homologies, the tree gets re-arranged due to the very low similarity percentage observed in the protein sequence between the first Mus musculus homologue (Flg from the mRNA homology page) and the Homo sapiens one, with only 22% as opposed to the 40% from the alignment of the other mouse protein sequence (against Homo sapien). This further suggests a possible misnomer in the identification of this gene as a homologue to the Homo sapiens filaggrin gene (and protein product). As shown in this tree, which displays the problematic mouse homologue at the very top, and the other one at the very bottom, we see a very early on split in the evolution of filaggrin amongst the species. Further research could reveal a very distinct function in this predicted Flg sequence that may distinguish it from its classification with these filaggrin genes and protein-products.
References
1. Homologene http://www.ncbi.nlm.nih.gov/homologene/
2. Wormbase http://www.wormbase.org/
3. Blast http://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ClustalWhttp://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=clustalw
5. Muscle http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=muscle
6. T-COFFEE http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=tcoffee
7. Gene Beehttp://www.genebee.msu.su/genebee.html
1. Homologene http://www.ncbi.nlm.nih.gov/homologene/
2. Wormbase http://www.wormbase.org/
3. Blast http://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ClustalWhttp://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=clustalw
5. Muscle http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=muscle
6. T-COFFEE http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=tcoffee
7. Gene Beehttp://www.genebee.msu.su/genebee.html